Data
This module has three data-related functions: (1) Datasets, (2) Offline Data Handling, and (3) Online Data Handling. The dataset functionality has built-in support for downloading and exploring a variety of validated datasets. The OfflineDataHandler is responsible for parsing through previously collected (offline) datasets and preparing data to be passed through the pipeline. Finally, the OnlineDataHandler is responsible for accumulating, storing, and processing real-time data.
Datasets
Several validated datasets consisting of different gestures and recording technology are included in this library. These datasets can be used for exploring the library’s capabilities and for future research. When using the packaged datasets for research purposes, we ask that you reference the original dataset contribution and not just this toolkit (the original dataset contributions might not be obvious since they are included for download with this toolkit).
Classification
OneSubjectMyoDataset
Dataset Description: Simple one subject dataset.
Attribute |
Description |
|---|---|
Num Subjects: |
1 |
Num Reps: |
12 Reps (i.e., 6 Trials x 2 Reps) |
Classes: |
|
Device: |
Myo |
Sampling Rates: |
200 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['OneSubjectMyo']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/OneSubjectEMaGerDataset
References:
@ARTICLE{libemg,
author={Eddy, Ethan and Campbell, Evan and Phinyomark, Angkoon and Bateman, Scott and Scheme, Erik},
journal={IEEE Access},
title={LibEMG: An Open Source Library to Facilitate the Exploration of Myoelectric Control},
year={2023},
volume={11},
number={},
pages={87380-87397},
keywords={Electromyography;Prosthetics;Libraries;Human computer interaction;Feature extraction;Muscles;Control systems;Gesture recognition;Open source software;EMG;electromyography;toolkit;library;myoelectric control;gesture recognition},
doi={10.1109/ACCESS.2023.3304544}}
EMGEPN100
Dataset Description: Multi-hardware EMG dataset for 12 different hand gesture categories using the Myo armband and the G-force armband.
Attribute |
Description |
|---|---|
Num Subjects: |
85 |
Num Reps: |
30 Reps x 12 Gestures x 43 Users (Train group), 15 Reps x 12 Gestures x 42 Users (Test group) –> Cross User Split |
Classes: |
|
Device: |
Myo, gForce |
Sampling Rates: |
Myo: 200Hz, gForce: 500Hz |
Auto Download: |
False |
Using the Dataset:
import libemg
from libemg.datasets import get_dataset_list
emg_epn100 = libemg.datasets.EMGEPN100()
# or get_dataset_list(cross_user=True)['EMGEPN100']()
odh = emg_epn100.prepare_data(split=True, segment=True, relabel_seg=None,
channel_last=True, subjects=None)['All']
Dataset Location https://laboratorio-ia.epn.edu.ec/es/recursos/dataset/emg-imu-epn-100
References:
@article{vasconez-2022,
author = {Vásconez, Juan Pablo and López, Lorena Isabel Barona and Caraguay, Ángel Leonardo Valdivieso and Benalcázar, Marco E.},
journal = {Sensors},
month = {12},
number = {24},
pages = {9613},
title = {{Hand Gesture Recognition Using EMG-IMU Signals and Deep Q-Networks}},
volume = {22},
year = {2022},
doi = {10.3390/s22249613},
url = {https://doi.org/10.3390/s22249613},
}
DS1: 3DC
Dataset Description: A relatively simple within session baseline.
Attribute |
Description |
|---|---|
Num Subjects: |
22 |
Num Reps: |
4 Training, 4 Testing |
Classes: |
|
Device: |
Delsys |
Sampling Rates: |
1000 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['3DC']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/3DCDataset
References:
@article{cote2019low,
title={A low-cost, wireless, 3-D-printed custom armband for sEMG hand gesture recognition},
author={C{\^o}t{\'e}-Allard, Ulysse and Gagnon-Turcotte, Gabriel and Laviolette, Fran{\c{c}}ois and Gosselin, Benoit},
journal={Sensors},
volume={19},
number={12},
pages={2811},
year={2019},
publisher={MDPI}
}
DS2: ContinuousTransitions
Dataset Description: The testing set in this dataset has continuous transitions between classes, providing a more realistic offline evaluation standard for myoelectric control.
Attribute |
Description |
|---|---|
Num Subjects: |
43 |
Num Reps: |
6 Training (Ramp), 42 Transitions (All combinations of Transitions) x 6 Reps |
Classes: |
|
Device: |
Delsys |
Sampling Rates: |
2000 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['ContinuousTransitions']()
odh = dataset.prepare_data()
Dataset Location https://unbcloud-my.sharepoint.com/:f:/g/personal/ecampbe2_unb_ca/EjgjhM9ZHJxOglKoAf062ngBf4wFj2Mn2bORKY1-aMYGRw?e=WkZNwI
References:
@ARTICLE{transitions,
author={Raghu, Shriram Tallam Puranam and MacIsaac, Dawn and Scheme, Erik},
journal={IEEE Journal of Biomedical and Health Informatics},
title={Decision-Change Informed Rejection Improves Robustness in Pattern Recognition-Based Myoelectric Control},
year={2023},
volume={27},
number={12},
pages={6051-6061},
doi={10.1109/JBHI.2023.3316599}}
DS3: Contraction Intensity
Dataset Description: A contraction intensity dataset.
Attribute |
Description |
|---|---|
Num Subjects: |
10 |
Num Reps: |
4 Ramp Reps (Train), 4 Reps x 20%, 30%, 40%, 50%, 60%, 70%, 80%, MVC (Test) |
Classes: |
|
Device: |
BE328 by Liberating Technologies, Inc |
Sampling Rates: |
1000 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['ContractionIntensity']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/ContractionIntensity
References:
@article{contraction_intensity,
title={Training strategies for mitigating the effect of proportional control on classification in pattern recognition--based myoelectric control},
author={Scheme, Erik and Englehart, Kevin},
journal={JPO: Journal of Prosthetics and Orthotics},
volume={25},
number={2},
pages={76--83},
year={2013},
publisher={LWW}
}
DS4: Electrode Shift
Dataset Description: An electrode shift confounding factors dataset.
Attribute |
Description |
|---|---|
Num Subjects: |
21 |
Num Reps: |
5 Train (Before Shift), 8 Test (After Shift) |
Classes: |
|
Device: |
Myo Armband |
Sampling Rates: |
200 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['ElectrodeShift']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/CIILData
References:
@article{ciil_es,
title={Context-informed incremental learning improves both the performance and resilience of myoelectric control},
author={Campbell, Evan and Eddy, Ethan and Bateman, Scott and C{\^o}t{\'e}-Allard, Ulysse and Scheme, Erik},
journal={Journal of NeuroEngineering and Rehabilitation},
volume={21},
number={1},
pages={70},
year={2024},
publisher={Springer}
}
DS5: EMGEPN612
Dataset Description: A large 612 user dataset for developing cross-user models.
Attribute |
Description |
|---|---|
Num Subjects: |
612 |
Num Reps: |
50 Reps x 306 Users (Train), 25 Reps x 306 Users (Test) –> Cross User Split |
Classes: |
|
Device: |
Myo Armband |
Sampling Rates: |
200 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['EMGEPN612']() # User Dependent
dataset = get_dataset_list(cross_user=True)['EMGEPN612']() # User Independent
odh = dataset.prepare_data()
Dataset Location https://unbcloud-my.sharepoint.com/:u:/g/personal/ecampbe2_unb_ca/EWf3sEvRxg9HuAmGoBG2vYkBLyFv6UrPYGwAISPDW9dBXw?e=vjCA14
References:
@article{epn,
title={EMG-EPN-612 Dataset. 2020},
author={Benalc{\'a}zar, M and Barona, L and Valdivieso, L and Aguas, X and Zea, J},
journal={DOI: https://doi. org/10.5281/zenodo},
volume={4027874},
year={2020}
}
DS6: FORSEMG
Dataset Description: Twelve gestures elicited in three forearm orientations (neutral, pronation, and supination).
Attribute |
Description |
|---|---|
Num Subjects: |
19 |
Num Reps: |
5 Train, 10 Test (2 Forearm Orientations x 5 Reps) |
Classes: |
|
Device: |
Experimental Device |
Sampling Rates: |
985 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['FORS-EMG']()
odh = dataset.prepare_data()
Dataset Location https://www.kaggle.com/datasets/ummerummanchaity/fors-emg-a-novel-semg-dataset
References:
@article{fors_emg,
title={FORS-EMG: A Novel sEMG Dataset for Hand Gesture Recognition Across Multiple Forearm Orientations},
author={Rumman, Umme and Ferdousi, Arifa and Hossain, Md Sazzad and Islam, Md Johirul and Ahmad, Shamim and Reaz, Mamun Bin Ibne and Islam, Md Rezaul},
journal={arXiv preprint arXiv:2409.07484},
year={2024}
}
DS7: Limb Position (Fougner)
Dataset Description: A limb position dataset (with 5 static limb positions).
Attribute |
Description |
|---|---|
Num Subjects: |
12 |
Num Reps: |
10 Reps (Train), 10 Reps x 4 Positions |
Classes: |
|
Device: |
BE328 by Liberating Technologies, Inc. |
Sampling Rates: |
1000 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['FougnerLP']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/LimbPosition
References:
@article{fougner_lp,
title={Resolving the limb position effect in myoelectric pattern recognition},
author={Fougner, Anders and Scheme, Erik and Chan, Adrian DC and Englehart, Kevin and Stavdahl, {\O}yvind},
journal={IEEE Transactions on Neural Systems and Rehabilitation Engineering},
volume={19},
number={6},
pages={644--651},
year={2011},
publisher={IEEE}
}
DS8: GRABMyo
Dataset Description: A large cross-session dataset including 17 gestures elicited across 3 separate sessions.
Attribute |
Description |
|---|---|
Num Subjects: |
43 |
Num Reps: |
7 Train, 14 Test (2 Separate Days x 7 Reps) –> Cross Day Split |
Classes: |
|
Device: |
EMGUSB2+ device (OT Bioelletronica, Italy) |
Sampling Rates: |
2048 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['GRABMyoBaseline']() # Baseline
dataset = get_dataset_list()['GRABMyoCrossDay']() # CrossDay
odh = dataset.prepare_data()
Dataset Location https://physionet.org/content/grabmyo/1.0.2/
References:
@article{grabmyo,
title={Multi-day dataset of forearm and wrist electromyogram for hand gesture recognition and biometrics},
author={Pradhan, Ashirbad and He, Jiayuan and Jiang, Ning},
journal={Scientific data},
volume={9},
number={1},
pages={733},
year={2022},
publisher={Nature Publishing Group UK London}
}
DS9: HyserPR
Dataset Description: High Density Hyser pattern recognition (PR) dataset. Includes dynamic and maintenance tasks for 34 hand gestures.
Attribute |
Description |
|---|---|
Num Subjects: |
18 |
Num Reps: |
1 Train, 1 Test (Consisting of dynamic and maintanance tasks) |
Classes: |
|
Device: |
OT Bioelettronica Quattrocento |
Sampling Rates: |
2048 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['HyserPR']()
odh = dataset.prepare_data()
Dataset Location https://www.physionet.org/content/hd-semg/2.0.0/
References:
@ARTICLE{hyser,
author={Jiang, Xinyu and Liu, Xiangyu and Fan, Jiahao and Ye, Xinming and Dai, Chenyun and Clancy, Edward A. and Akay, Metin and Chen, Wei},
journal={IEEE Transactions on Neural Systems and Rehabilitation Engineering},
title={Open Access Dataset, Toolbox and Benchmark Processing Results of High-Density Surface Electromyogram Recordings},
year={2021},
volume={29},
number={},
pages={1035-1046},
doi={10.1109/TNSRE.2021.3082551}}
DS10: Single-User Multi-Day (KaufmannMD)
Dataset Description: A single subject, multi-day (120 days) collection.
Attribute |
Description |
|---|---|
Num Subjects: |
1 |
Num Reps: |
1 rep per day, 120 days total. 60/60 train-test split |
Classes: |
|
Device: |
MindMedia |
Sampling Rates: |
2048 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['KaufmannMD']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/MultiDay
References:
@INPROCEEDINGS{kaufmann,
author={Kaufmann, Paul and Englehart, Kevin and Platzner, Marco},
booktitle={2010 Annual International Conference of the IEEE Engineering in Medicine and Biology},
title={Fluctuating emg signals: Investigating long-term effects of pattern matching algorithms},
year={2010},
volume={},
number={},
pages={6357-6360},
doi={10.1109/IEMBS.2010.5627288}}
DS11: Minimal Training Data
Dataset Description: The goal of this Myo dataset is to explore how well models perform when they have a limited amount of training data (1s per class).
Attribute |
Description |
|---|---|
Num Subjects: |
11 |
Num Reps: |
1 Train, 15 Test |
Classes: |
|
Device: |
Myo Armband |
Sampling Rates: |
200 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['MinimalTrainingData']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/CIILData
References:
@inproceedings{ciil_md,
title={Leveraging task-specific context to improve unsupervised adaptation for myoelectric control},
author={Eddy, Ethan and Campbell, Evan and Bateman, Scott and Scheme, Erik},
booktitle={2023 IEEE International Conference on Systems, Man, and Cybernetics (SMC)},
pages={4661--4666},
year={2023},
organization={IEEE}
}
DS12: NinaProDB2
Dataset Description: The Ninapro DB2 is a dataset that can be used to test how algorithms perform for large gesture sets. The dataset contains 6 repetitions of 50 motion classes (plus optional rest) that were recorded using 12 Delsys Trigno electrodes around the forearm.
Attribute |
Description |
|---|---|
Num Subjects: |
40 |
Num Reps: |
6 |
Classes: |
50 Nina Pro DB2 |
Device: |
Delsys |
Sampling Rates: |
2000 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['NinaProDB2']()
odh = dataset.prepare_data()
Dataset Location Note, this dataset will not be automatically downloaded. To download this dataset, please see Nina DB2. Simply download the ZIPs and place them in a folder and LibEMG will handle the rest. All credit for this dataset should be given to the original authors.
References:
@article{db2,
title={Electromyography data for non-invasive naturally-controlled robotic hand prostheses},
author={Atzori, Manfredo and Gijsberts, Arjan and Castellini, Claudio and Caputo, Barbara and Hager, Anne-Gabrielle Mittaz and Elsig, Simone and Giatsidis, Giorgio and Bassetto, Franco and M{\"u}ller, Henning},
journal={Scientific data},
volume={1},
number={1},
pages={1--13},
year={2014},
publisher={Nature Publishing Group}
}
DS13: Limb Position (Radmand)
Dataset Description: A large limb position dataset (with 16 static limb positions).
Attribute |
Description |
|---|---|
Num Subjects: |
10 |
Num Reps: |
4 Reps (Train), 4 Reps x 15 Positions |
Classes: |
|
Device: |
DelsysTrigno |
Sampling Rates: |
1000 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list()['RadmandLP']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/LimbPosition
References:
@INPROCEEDINGS{radmand_lp,
author={Radmand, A. and Scheme, E. and Englehart, K.},
booktitle={2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society},
title={A characterization of the effect of limb position on EMG features to guide the development of effective prosthetic control schemes},
year={2014},
volume={},
number={},
pages={662-667},
keywords={},
doi={10.1109/EMBC.2014.6943678}}
DS14: Pre/Post TMR
Dataset Description: 6 subjects, 8 reps, 24 motions, pre/post intervention.
Attribute |
Description |
|---|---|
Num Subjects: |
6 |
Num Reps: |
8 reps per motion (pre/post intervention) |
Classes: |
|
Device: |
Ag/AgCl |
Sampling Rates: |
1000 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset_pre = get_dataset_list()['TMR_Pre']()
dataset_post = get_dataset_list()['TMR_Post']()
odh = dataset_pre.prepare_data()
Dataset Location https://github.com/LibEMG/TMR_ShirleyRyanAbilityLab
References:
@article{tmr,
title={Myoelectric prosthesis hand grasp control following targeted muscle reinnervation in individuals with transradial amputation},
author={Simon, Ann M and Turner, Kristi L and Miller, Laura A and Dumanian, Gregory A and Potter, Benjamin K and Beachler, Mark D and Hargrove, Levi J and Kuiken, Todd A},
journal={PloS one},
volume={18},
number={1},
pages={e0280210},
year={2023},
publisher={Public Library of Science San Francisco, CA USA}
}
DS15: Weakly Supervised
Dataset Description: A weakly supervised environment with sparse supervised calibration.
Attribute |
Description |
|---|---|
Num Subjects: |
16 |
Num Reps: |
30 min weakly supervised, 1 rep calibration, 14 reps test |
Classes: |
|
Device: |
OyMotion gForcePro+ EMG Armband |
Sampling Rates: |
1000 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list('WEAKLYSUPERVISED')['WeaklySupervised']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/WS_CIIL
References:
@ARTICLE{10802919,
author={Campbell, Evan and Eddy, Ethan and Isabel, Xavier and Bateman, Scott and Gosselin, Benoit and Côté-Allard, Ulysse and Scheme, Erik},
journal={IEEE Transactions on Neural Systems and Rehabilitation Engineering},
title={Screen Guided Training Does Not Capture Goal-Oriented Behaviors: Learning Myoelectric Control Mappings From Scratch Using Context Informed Incremental Learning},
year={2025},
volume={33},
number={},
pages={332-342},
keywords={Training;Electromyography;Games;Data models;Calibration;Adaptation models;Context modeling;Predictive models;Pipelines;Wrist;Electromyography (EMG);deep learning;incremental learning;gesture recognition;zero-shot},
doi={10.1109/TNSRE.2024.3518059}}
Regression
OneSubjectEmaGEr
Dataset Description: Simple one subject regression dataset.
Attribute |
Description |
|---|---|
Num Subjects: |
1 |
Num Reps: |
5 Reps |
Classes: |
|
Device: |
EmaGEr |
Sampling Rates: |
1010 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list('REGRESSION')['OneSubjectEMaGer']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/OneSubjectEMaGerDataset
References:
@ARTICLE{libemg,
author={Eddy, Ethan and Campbell, Evan and Phinyomark, Angkoon and Bateman, Scott and Scheme, Erik},
journal={IEEE Access},
title={LibEMG: An Open Source Library to Facilitate the Exploration of Myoelectric Control},
year={2023},
volume={11},
number={},
pages={87380-87397},
doi={10.1109/ACCESS.2023.3304544}}
DS16: EMG2POSE
Dataset Description: A large dataset from ctrl-labs (Meta) for joint angle estimation. Note that not all subjects have all stages.
Attribute |
Description |
|---|---|
Num Subjects: |
193 |
Num Reps: |
N/A |
Classes: |
|
Device: |
Ctrl Labs Armband |
Sampling Rates: |
2000 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list('REGRESSION')['EMG2POSE']() # Within USer
dataset = get_dataset_list('REGRESSION', cross_user=True)['EMG2POSE']() # Cross User
odh = dataset.prepare_data()
Dataset Location https://fb-ctrl-oss.s3.amazonaws.com/emg2pose/emg2pose_dataset.tar
References:
@inproceedings{salteremg2pose,
title={emg2pose: A Large and Diverse Benchmark for Surface Electromyographic Hand Pose Estimation},
author={Salter, Sasha and Warren, Richard and Schlager, Collin and Spurr, Adrian and Han, Shangchen and Bhasin, Rohin and Cai, Yujun and Walkington, Peter and Bolarinwa, Anuoluwapo and Wang, Robert and others},
booktitle={The Thirty-eight Conference on Neural Information Processing Systems Datasets and Benchmarks Track}
}
DS17: Hyser1DOF
Dataset Description:
Hyser 1 DOF dataset. Includes within-DOF finger movements. Ground truth finger forces are recorded for use in finger force regression.
Attribute |
Description |
|---|---|
Num Subjects: |
20 |
Num Reps: |
3 |
Classes: |
|
Device: |
OT Bioelettronica Quattrocento |
Sampling Rates: |
2048 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list('REGRESSION')['Hyser1DOF']()
odh = dataset.prepare_data()
Dataset Location https://www.physionet.org/content/hd-semg/2.0.0/
References:
@ARTICLE{hyser,
author={Jiang, Xinyu and Liu, Xiangyu and Fan, Jiahao and Ye, Xinming and Dai, Chenyun and Clancy, Edward A. and Akay, Metin and Chen, Wei},
journal={IEEE Transactions on Neural Systems and Rehabilitation Engineering},
title={Open Access Dataset, Toolbox and Benchmark Processing Results of High-Density Surface Electromyogram Recordings},
year={2021},
volume={29},
number={},
pages={1035-1046},
doi={10.1109/TNSRE.2021.3082551}}
DS18: HyserNDOF
Dataset Description:
Hyser N DOF dataset. Includes combined finger movements. Ground truth finger forces are recorded for use in finger force regression.
Attribute |
Description |
|---|---|
Num Subjects: |
20 |
Num Reps: |
2 |
Classes: |
|
Device: |
OT Bioelettronica Quattrocento |
Sampling Rates: |
2048 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list('REGRESSION')['HyserNDOF']()
odh = dataset.prepare_data()
Dataset Location https://www.physionet.org/content/hd-semg/2.0.0/
References:
@ARTICLE{hyser,
author={Jiang, Xinyu and Liu, Xiangyu and Fan, Jiahao and Ye, Xinming and Dai, Chenyun and Clancy, Edward A. and Akay, Metin and Chen, Wei},
journal={IEEE Transactions on Neural Systems and Rehabilitation Engineering},
title={Open Access Dataset, Toolbox and Benchmark Processing Results of High-Density Surface Electromyogram Recordings},
year={2021},
volume={29},
number={},
pages={1035-1046},
doi={10.1109/TNSRE.2021.3082551}}
DS19: HyserRandom
Dataset Description:
Hyser random dataset. Includes random motions performed by users. Ground truth finger forces are recorded for use in finger force regression.
Attribute |
Description |
|---|---|
Num Subjects: |
19 |
Num Reps: |
5 |
Classes: |
Random |
Device: |
OT Bioelettronica Quattrocento |
Sampling Rates: |
2048 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list('REGRESSION')['HyserRandom']()
odh = dataset.prepare_data()
Dataset Location https://www.physionet.org/content/hd-semg/2.0.0/
References:
@ARTICLE{hyser,
author={Jiang, Xinyu and Liu, Xiangyu and Fan, Jiahao and Ye, Xinming and Dai, Chenyun and Clancy, Edward A. and Akay, Metin and Chen, Wei},
journal={IEEE Transactions on Neural Systems and Rehabilitation Engineering},
title={Open Access Dataset, Toolbox and Benchmark Processing Results of High-Density Surface Electromyogram Recordings},
year={2021},
volume={29},
number={},
pages={1035-1046},
doi={10.1109/TNSRE.2021.3082551}}
DS20: NinaProDB8
Attribute |
Description |
|---|---|
Num Subjects: |
12 |
Num Reps: |
20 Training, 2 Testing |
Classes: |
|
Device: |
Delsys |
Sampling Rates: |
1111 Hz |
Auto Download: |
False |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list('REGRESSION')['NinaProDB8']()
odh = dataset.prepare_data()
Dataset Location Note, this dataset will not be automatically downloaded. To download this dataset, please see Nina DB8. Simply download the ZIPs and place them in a folder and LibEMG will handle the rest. All credit for this dataset should be given to the original authors.
References:
@article{db8,
title={Effect of user practice on prosthetic finger control with an intuitive myoelectric decoder},
author={Krasoulis, Agamemnon and Vijayakumar, Sethu and Nazarpour, Kianoush},
journal={Frontiers in neuroscience},
volume={13},
pages={891},
year={2019},
publisher={Frontiers Media SA}
}
DS21: UserCompliance
Dataset Description:
Regression dataset used for investigation into user compliance during mimic training.
Attribute |
Description |
|---|---|
Num Subjects: |
6 |
Num Reps: |
5 |
Classes: |
|
Device: |
EMaGer |
Sampling Rates: |
1010 Hz |
Auto Download: |
True |
Using the Dataset:
from libemg.datasets import *
dataset = get_dataset_list('REGRESSION')['UserCompliance']()
odh = dataset.prepare_data()
Dataset Location https://github.com/LibEMG/UserComplianceDataset
References:
@inproceedings{morrell2024exploring,
title={Exploring user compliance in the training of regression-based myoelectric control},
author={Morrell, Christian and Campbell, Evan and Scheme, Erik},
booktitle={Myoelectric Controls Symposium},
year={2024}
}
Offline Data Handler
One overhead for most EMG projects is interfacing with a particular dataset since they often have different folder and file structures. LibEMG provides a means to quickly interface datasets so you can focus on using them with minimal setup time. Assuming the files in the dataset are well formatted (i.e., they include all metadata such as rep, class, and subject) and are either .csv or .txt files, the OfflineDataHandler does all accumulation and processing. To do this, LibEMG relies on regular expressions to define a dataset’s file and folder structure. These expressions can be used to create a dictionary that is passed to the OfflineDataHandler. Once the data handler has collected all the files that satisfy the regexes, the dataset can be sliced using the metadata tags (e.g., by rep, subjects, classes, etc.). After extracting the data it is ready to be passed through the rest of the pipeline. The following code snippet exemplifies how to process a dataset with testing/training, rep, and class metadata. In this case the file format is: dataset/train/R_1_C_1_EMG.csv where R is the rep and C is the class.
Example Code
from libemg.data_handler import OfflineDataHandler, RegexFilter
dataset_folder = 'dataset'
regex_filters = [
RegexFilter(left_bound = "dataset/", right_bound="/", values = sets_values, description='sets'),
RegexFilter(left_bound = "_C_", right_bound="_EMG.csv", values = classes_values, description='classes'),
RegexFilter(left_bound = "R_", right_bound="_C_", values = reps_values, description='reps')
]
odh = OfflineDataHandler()
odh.get_data(folder_location=dataset_folder, regex_filters=regex_filters, delimiter=",")
# Extract training data:
train_odh = odh.isolate_data(key="sets", values=[0])
train_windows, train_metadata = train_odh.parse_windows(50,25)
# Extract features
fe = FeatureExtractor()
feature_list = fe.get_feature_list()
training_features = fe.extract_features(feature_list, train_windows)
Online Data Handler
One complication when using EMG devices is the lack of standardization, meaning that interfacing with hardware is a new undertaking for each device. A goal of LibEMG is to abstract these differences and enable a hardware-agnostic framework. Therefore, this module acts as a middle layer for processing real-time data streaming from any device. In this architecture - exemplified in Figure 1 – live data streaming is performed by using a shared memory buffer as the core. This shared memory buffer is created by the device streamer, where a process is spawned that continuously populates the buffer with samples. Other modules can gain access to the shared memory buffer using the shared memory items that the streamer returned, allowing for cross-process, low-latency, non-blocking access to the data of interest. We provide an OnlineDataHandler object that is a generic object for attaching to the shared memory buffer with added some utilities.
An example of the online data streaming workflow is provided below:
# start the hardware streamer, receive a process handle and shared memory descriptors
streamer_process, shared_memory_items = libemg.streamers.myo_streamer()
# start an online data handler to attach to the shared memory buffer
odh = libemg.data_handler.OnlineDataHandler(shared_memory_items=shared_memory_items)
# start logging the data to a file
odh.log_to_file()
# start visualizing the data
odh.visualization()
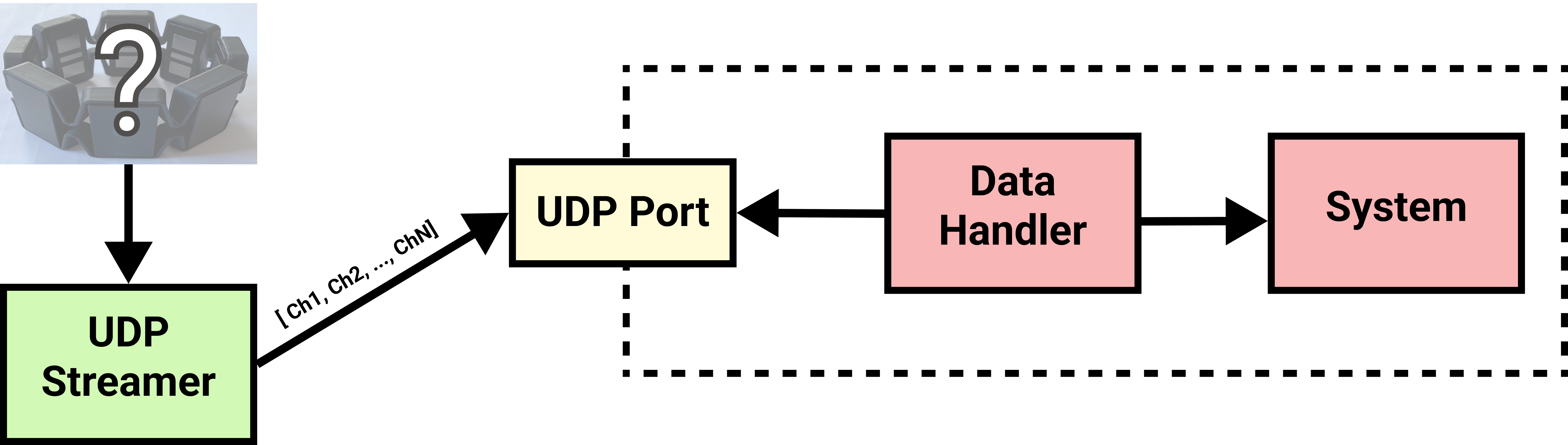
Figure 1: OnlineDataHandler Architecture
For more information on the default streamers and creating your own, please reference the Supported Hardware section.